Mais informações
Bem-vindo ao blog do Laboratório de Bioinformática Estrutural (LaBiE) da ULBRA
Localização:
Av. Farroupilha, 8001 - Prédio 01, Sala 122 - Bairro São José, Canoas/RS
CEP: 92425-900 - Tel: +55(51)34774000 ext 2774
Últimas notícias em Bioinf & QM - links e posts
quarta-feira, 4 de abril de 2012
II Escola e Bioinformática Estrutural da Ulbra
Mais informações
domingo, 1 de abril de 2012
Exemplo de estudo brasileiro coordenado
Synthesis, Biological Evaluation, And Molecular Modeling of Chalcone Derivatives As Potent Inhibitors of Mycobacterium tuberculosis Protein Tyrosine Phosphatases (PtpA and PtpB)
sexta-feira, 2 de setembro de 2011
terça-feira, 23 de agosto de 2011
Toxic antibodies blitz tumours
Tightly targeted cancer therapy receives marketing approval
Eventually, Clay Siegall got used to doors slamming in his face. When the cancer researcher decided to create a company that would fight cancer using weaponized antibodies, investors were sceptical. "We contacted 35, then 40, then 45 venture-capital companies," he says. "We got turned down over and over and over."
Siegall and his partners kept trying, and 13 years later the investors who eventually bet on Siegall's company, Seattle Genetics, of Bothell, Washington, are getting their reward. On 19 August, the US Food and Drug Administration (FDA) approved the company's lead therapy, an antibody engineered to deliver a poisonous payload directly into lymphoma cells. The hope is that such antibodies, called antibody–drug conjugates, will sidestep the punishing toxicities of classical chemotherapies, which run loose in the bloodstream and kill healthy cells in addition to their targets.
 Click for full graphic
Click for full graphicTim Illidge, an oncologist at the University of Manchester, UK, says that the approval is a "game-changer" for a promising class of drugs that has struggled to gain a foothold since it was first described in a 1964 Nature paper1 (see 'A long time coming'). "We're essentially in a renaissance of the antibody–drug conjugate," he says.
Unembellished, or 'naked', antibodies are already used to treat cancer because of their unparallelled ability to target proteins found on the surface of tumour cells. Their high profit margins and strong patent protections have pharmaceutical companies clamouring for more. Siegall says that Seattle Genetics toyed with naked antibodies, too. "But by and large," he says, "most naked antibodies just don't have a strong, potent ability to knock out tumour cells."
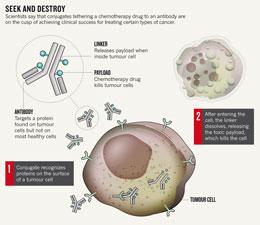
Enter antibody–drug conjugates, which do have that knock-out punch. Their power comes from their payloads: lethal drugs tethered to the antibody that remain harmless until the conjugate releases them inside cancer cells (see 'Seek and destroy').
But for decades, developers have struggled to get the crucial elements to work together: the antibody, the drug it carries and the linker that binds the two. Seattle Genetics' conjugate, Adcetris (brentuximab vedotin), seems to have overcome the hurdles; it combines a synthetic poison called vedotin with an antibody that targets CD30, a protein found on many lymphoma cells.
In July, an FDA advisory committee voted unanimously in support of accelerated approval for Adcetris after Seattle Genetics reported that 94% of the 102 people with Hodgkin's lymphoma in a trial of the treatment saw their tumours shrink, and 73% achieved partial or complete remission. "This drug is wildly active," said panellist Mikkael Sekeres, an oncologist at the Cleveland Clinic in Ohio, after the vote.
The accelerated approval means that the drug can now be prescribed by doctors while Seattle Genetics conducts follow-up clinical studies. Mark Monane, a senior analyst at the investment-banking firm Needham & Company in New York, predicts that the drug will bring in up to US$400 million a year in sales.
“It’s like a game of whack-a-mole. You knock out one toxicity and another shows up.”
Janice Reichert, an analyst at the Tufts Center for the Study of Drug Development in Boston, Massachusetts, expects more approvals to follow. Between 2000 and 2005, only six antibody–drug conjugates entered the clinic for the first time, she says. From 2005 to 2009, 15 more joined their ranks. Now, 25 are currently in cancer clinical trials — more than at any other time. Two have reached late-stage clinical trials: trastuzumab emtansine, a breast-cancer therapy jointly developed by biotechnology firms Genentech, based in South San Francisco, and ImmunoGen in Waltham, Massachusetts; and inotuzumab ozogamicin, a lymphoma therapy developed by Pfizer in New York.
Pitfalls remain. The only other conjugate to ever win accelerated approval from the FDA, a different version of Pfizer's lymphoma therapy, was pulled from the market last year after further tests showed that the drug offered no benefits over standard chemotherapy. Many blame the failure on a linker that fell apart in the bloodstream, boosting toxicity and limiting the dose that could be used. Seattle Genetics had to pull one of its antibodies from clinical trials for similar reasons, says Siegall. As a result, the company developed a linker that is degraded by the enzymes that are most active inside the cell.
ImmunoGen, Seattle Genetics' main competitor, has struggled to find the right drug to couple to its antibodies. For years, the company attempted to use ricin, a toxin produced by castor beans. But ricin triggered a dangerous immune response. ImmunoGen now steers clear of complex proteins and picks small-molecule poisons that are less likely to attract the attention of the immune system.
A lingering problem for the field is a lack of control over how many drug molecules attach to each antibody. More control would help standardize each dose and lessen the potential toxicity of the treatment. A method developed by researchers at Genentech, a subsidiary of Swiss drug firm Roche, seemed to have conquered the problem but has not yet been tested clinically2. Unexpected toxicities led the company to shelve the technique for the time being, says Paul Polakis, Genentech's director of cancer targets. "It's like a game of whack-a-mole," he sighs. "You knock out one toxicity and another shows up."
Although antibody engineers still have work to do in optimizing the design, the approval of Adcetris means they have a bellwether to watch. The improving fortunes in the field, which culminated in last week's approval, have finally brought Siegall the investor attention that eluded him for so long. Seattle Genetics has partnered with 11 outside firms, bringing in $150 million in new capital. More than a quarter of that was raised in the past year. "I'm happy to say I no longer have to convince investors that this is a productive field," he says.
All of this leaves those who have followed the technology marvelling at its reversal of fortune. "When I first became interested in the field in the early 1990s, there was a lot of despondency," says Illidge. "And now look at it. Everything has changed."
sexta-feira, 19 de agosto de 2011
Fighting neurodegeneration with rapamycin: mechanistic insights
Jordi Bové, Marta Martínez-Vicente & Miquel Vila
Abstract
A growing number of studies point to rapamycin as a pharmacological compound that is able to provide neuroprotection in several experimental models of neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, Huntington's disease and spinocerebellar ataxia type 3. In addition, rapamycin exerts strong anti-ageing effects in several species, including mammals. By inhibiting the activity of mammalian target of rapamycin (mTOR), rapamycin influences a variety of essential cellular processes, such as cell growth and proliferation, protein synthesis and autophagy. Here, we review the molecular mechanisms underlying the neuroprotective effects of rapamycin and discuss the therapeutic potential of this compound for neurodegenerative diseases.
http://www.nature.com/nrn/journal/v12/n8/full/nrn3068.html
domingo, 10 de julho de 2011

Acontecerá nos dias 12-15 de outubro na cidade de Floránopolis - SC o 7th International Conference of the Brazilian Association for Bioinformatics and Computational Biology (AB3C) e o 3rd International Conference of the IberoAmerican Society for Bioinformatics (SoIBio). X-meetig 2011 (AB3C / SolBio).
O encontro contará com os palestrantes brasileiros Ana Tereza Vasconcelos (LNCC) e Aristóteles Góes-Neto (UEFS), bem como os palestrantes internacionais Ana Teresa Freitas (INESC Lisboa), Robin Haw (Ontario Inst. Cancer. Res.), Erik Lindahl (Stockholm University), Arthur J. Olson (Scripps Res. Inst.), entre outros.
As inscrições serão no período de 12 de setembro a 15 de outubro de 2011 no site http://www.x-meeting.com/ . A submissão de artigos será no período de 21 de junho a 1 de setembro de 2001 no site. Para maiores informações visite o site do x-meeting.
sexta-feira, 1 de abril de 2011
Landmark lupus approval opens door for next wave of drugs
Nature Reviews Drug Discovery 10, 243-245 (April 2011) | doi:10.1038/nrd3413
The US approval of Human Genome Sciences (HGS) and GlaxoSmithKline's belimumab (Benlysta) on 9 March ended a 50-year drought in the introduction of new lupus drugs. The two firms have blazed a path from mRNA to medicine in 15 years, and analysts are now predicting peak US sales of over US$2 billion annually for the monoclonal antibody (mAb).






